Hadrien Croubois: Toward an autonomic engine for scientific workflows and elastic Cloud infrastructure
Everyone is welcome to attend Hadrien Croubois’s thesis defense, which will take place Tuesday 16th October at 14h at Salle des thèse (ENS de Lyon).
You are also invited to the cocktail that follows the defense.
Advisors:
Eddy Caron, ENS de Lyon
Committee members:
Noël De Palma, Université Jospeh Fourier, reviewer
Johan Montagnat, CNRS, Laboratoire I3S UMR 7271, reviewer
Luciana Arantes, Université Sorbonne, examiner
Frédéric Desprez, Inria, examiner
Pushpinder Kaur Chouhan,Ulster University, examiner
Abstract:
The constant development of scientific and industrial computation infrastructures requires the concurrent development of scheduling and deployment mechanisms to manage such infrastructures. Throughout the last decade, the emergence of the Cloud paradigm raised many hopes, but achieving full platform autonomicity is still an ongoing challenge.
Work undertaken during this Ph.D. aimed at building a workflow engine that integrated the logic needed to manage workflow execution and Cloud deployment on its own. More precisely, we focus on Cloud solutions with a dedicated Data as a Service (DaaS) data management component. Our objective was to automate the execution of workflows submitted by many users on elastic Cloud resources.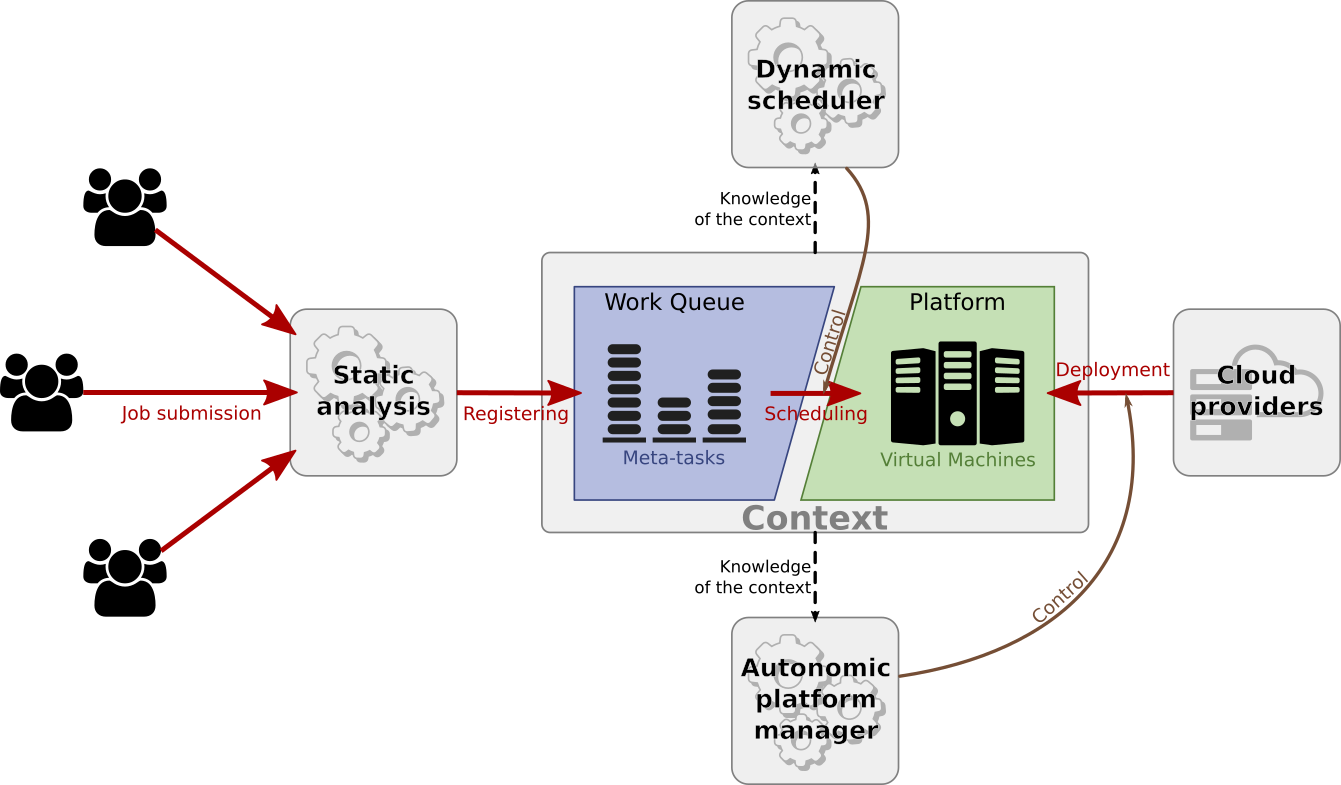
This contribution proposes a modular middleware infrastructure and details the implementation of the underlying modules:
- A workflow clustering algorithm that optimises data locality in the context of DaaS-centered communications;
- A dynamic scheduler that executes clustered workflows on Cloud resources;
- A deployment manager that handles the allocation and deallocation of Cloud resources according to the workload characteristics and users’ requirements.
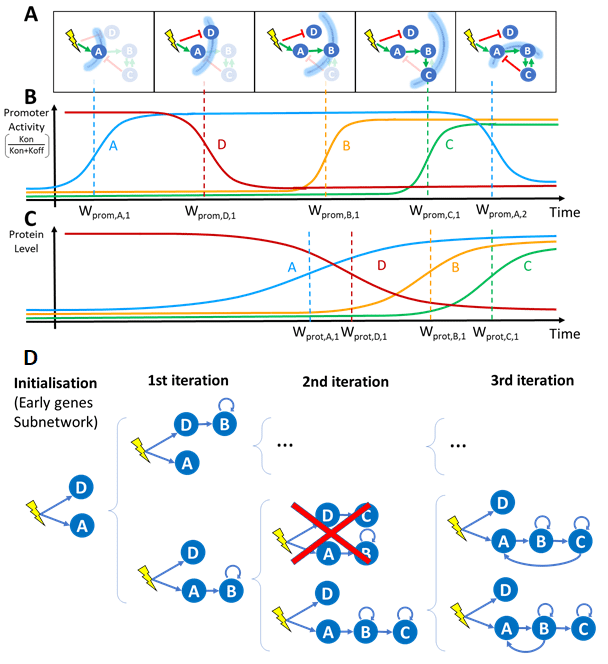
All these modules have been implemented in a simulator to analyse their behaviour and measure their effectiveness when running both synthetic and real scientific workflows. We also implemented these modules in the DIET middleware to give it new features and prove the versatility of this approach. Simulation running the WASABI workflow (waves analysis based inference, a framework for the reconstruction of gene regulatory networks) showed that our approach can decrease the deployment cost by up to 44% while meeting the required deadlines.



 Le premier ministre Édouard Philippe a annoncé le 1er février 2019, le lancement d’une loi de programmation pluriannuelle pour la recherche à l’occasion des 80 ans du CNRS à la Maison de la Mutualité, à Paris.
Le premier ministre Édouard Philippe a annoncé le 1er février 2019, le lancement d’une loi de programmation pluriannuelle pour la recherche à l’occasion des 80 ans du CNRS à la Maison de la Mutualité, à Paris.
 Plongez dans l’univers du Numérique.
Plongez dans l’univers du Numérique.